Mar 09, 2022
AI for Simulation: how Graphcore is helping transform traditional HPC
Written By:
Alex Titterton

Mar 09, 2022
Written By:
Alex Titterton
Join the IPU conversation
Join our Graphcore community for free. Get help and share knowledge, find tutorials and tools that will help you grow.
Join on SlackFor many years High Performance Computing (HPC) techniques have been used to solve the world’s most complex scientific problems across a wide range of applications, from using Monte-Carlo simulation to model Higgs boson decay at the Large Hadron Collider to predicting whether the weather will improve.
However, due to the immense complexity of the calculations involved in many of these applications, researchers are often waiting a long time for simulation results to arrive. Speeding up these workflows by simply running the same programs on more powerful hardware can be very expensive, with a large cost often giving only a modest improvement in performance.
Clearly, a new approach is required to efficiently speed up these workloads, and many researchers are turning to surrogate machine learning models.
A surrogate model is a machine learning model intended to imitate part of a traditional HPC workflow, providing results in an accelerated time frame. Shown in Figure 1, this scheme is intended to replace the computationally intensive bottlenecks with machine learning-driven techniques, without needing to port the entire end-to-end workflow.
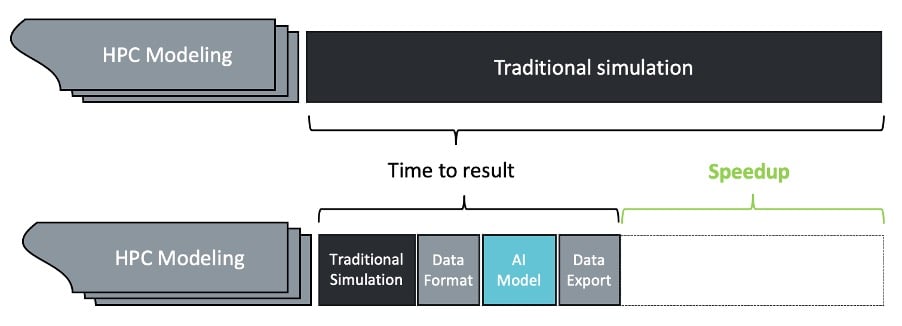
Figure 1: High-level schema for HPC speed-up with AI
For example, given a physical system, whose behaviour is to be simulated, a surrogate model can be used to approximate the results achieved by more computationally expensive Monte-Carlo simulation techniques, in a small fraction of the time. An AI-based solver can be used to give approximate solutions to sets of differential equations more quickly than can be obtained using numerical methods.
In cases where the data generated using traditional HPC methods is highly accurate, and the goal is to attain similar results more quickly, the machine learning models can be trained directly on simulated data. In many cases, machine learning-based approaches can even outperform traditional HPC emulation techniques in terms of accuracy, giving researchers the best of both worlds: more accurate results in a shorter time.
Graphcore’s Bow Pod systems, powered by the Intelligence Processing Unit (IPU), have been designed from the ground up to accelerate machine learning workloads, with plug-and-play connectivity for efficient datacentre scalability. Furthermore, Bow Pod's disaggregated architecture means the CPUs reside separately from the IPUs, meaning the CPU:IPU ratio can be varied depending on the workload requirements.
This flexible CPU:IPU ratio contributes to make the Bow Pods the ideal platform for accelerating HPC workloads using AI surrogate models; allowing researchers to accelerate the machine learning-based aspects of their workloads without hindering parts which may rely on more traditional CPU-workload HPC techniques.
In the next sections, we will explore a variety of applications that use AI to speed up parts of their HPC workload. All examples, unless otherwise stated, were run on second generation Graphcore IPU systems, prior to the launch of IPU Bow and the current Bow Pod range.
In a recent paper by M. Chantry et al, the European Centre for Medium-range Weather Forecasting (ECMWF) explored the use of deep learning models to emulate the effects of gravity wave drag in numerical weather forecasting. As part of this work, a range of deep learning models were designed to emulate the results of more traditional non-ML-based parametrisation schemes.
Chantry et al focused specifically on parameterising the effects of Non-Orographic Gravity Wave Drag (NOGWD), which is a zonal acceleration caused by the breaking of upward-propagating gravity waves, typically leading to turbulence and dissipation within the atmosphere. These types of gravity waves occur on widely varying scales, meaning that current forecast resolutions can resolve some but not all gravity waves. The effects of NOGWD on weather systems can occur over seasonal timescales, making effective parameterisation of this scheme important for medium to long-range weather forecasting.
Multi-Layer Perceptron (MLP) and Convolutional Neural Network (CNN) models were found to be more accurate for medium-range weather forecasting compared with the parametrisation schemes they were designed to emulate. However, training these models on a CPU gave no real speedup compared with traditional parametrisation methods, motivating the use of dedicated AI hardware.
Engineers at ATOS provided Graphcore with implementations of the MLP and CNN models in TensorFlow 2. These were ported to the IPU, requiring just a handful of code changes to train efficiently and successfully. Without any changes to the model definition or its hyperparameters, a single IPU processor was able to train this MLP model 5x faster than an A100 GPU and some 50x faster than either the MLP model or the traditional parametrisation scheme performed on a CPU. More information can be found in our weather forecasting blog post.
Deep learning approaches are being applied more and more within the field of Computational Fluid Dynamics (CFD). In many cases, problems are characterised by physical models defined by deterministic equations, meaning a data-driven approach can at best lead to approximate solutions. These problems can, however, often be broken down into sub-problems, each of which relies on a simpler set of assumptions rather than complex, well-defined equations. Data-driven machine learning approaches can be trained on data generated using more costly numerical simulations, and be used to provide an accurate model of the underlying system.
One such approach is the use of deep learning models in estimating the Sub-Grid Scale (SGS) contribution to flame surface density estimation in combustion modelling. In Large-Eddy Simulation (LES) a cut-off scale is defined such that reactions taking place on a smaller scale than is directly resolved are grouped into an estimated SGS contribution term, resulting in a trade-off between simulation accuracy and computational complexity.
Since the reliability of the LES model depends on accurate modelling of the SGS contribution but lowering the cut-off threshold would incur a large computational cost, the ability to model the SGS contribution, quickly and accurately, in flame surface density estimation would be highly valuable.
The European Center of Excellence in Exascale Computing "Research on AI- and Simulation-Based Engineering at Exascale" (CoE RAISE) investigated the use of a U-Net-inspired model for estimating the SGS contribution to the reaction rate for pre-mixed turbulent flames.
The model takes as input a 16 x 16 x 16 crop of a larger 64 x 32 x 32 3D volume representing the progress variable c, defined for a temperature T as:
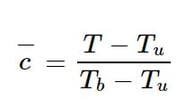
for each point, where the subscripts b and u refer to the temperatures of burnt and unburnt gases, respectively. The model output tensor approximates the flame surface density ∑ at each spatial point. Once trained, this model was found to outperform classical algebraic models to accurately estimate ∑.
Engineers at ATOS kindly provided Graphcore with an implementation of this U-Net inspired model, and with just a few code changes the model was ported to train on a single IPU processor. Furthermore, the model was found to train in around 20 minutes on a V100, and in around 5 minutes using a single IPU. In both cases the models converged in around 150 epochs, with the model trained on the IPU converging to a lower MSE loss.
Similarly to CFD modelling, efficient generation of simulated data in High-Energy Particle Physics (HEP) requires overcoming significant technical difficulties. One such challenge is accurate and fast simulation of particle “jets” in proton-proton collisions.
In a high-energy proton-proton collision, many subatomic particles are produced. According to the laws of Quantum Chromodynamics (QCD), certain types of produced particles such as quarks cannot exist freely and so hadronise, producing and bonding with other particles, resulting in narrow cone-shaped “jets”. The energies and momenta of these jets can then be measured in particle detectors to study the properties of the original quarks.
The simulation of jet production at the Large Hadron Collider (LHC) at CERN represents a large technical hurdle, due to the need to accurately simulate enormous amounts of particle collisions. To overcome this challenge, researchers have begun investigating the use of Generative Adversarial Networks (GANs) to generate realistic simulated data more quickly than can be achieved using traditional Monte-Carlo methods.
Typically, a GAN consists of two networks, a generator and a discriminator. The generator turns random noise into a data sample, and the discriminator attempts to distinguish between real and generated data samples, with each network being trained based on the outputs of the other. GANs have long been able to generate photo-realistic images and are being used increasingly in particle physics applications, both for generating simulated collision data and to accurately model the physical characteristics of a particle detector.
Researchers at the University of Bristol, working on the LHCb experiment at the LHC, compared GAN training and inference performance on Graphcore IPUs and existing GPU-based solutions. In this case a CNN-based DiJetGAN model was trained to simulate the production of particle jets at the LHC.
It was found that a single GC2 (1st generation) IPU processor was able to deliver a performance gain of up to 5.4x over a P100 GPU, consuming only half the power. With the latest, flexible Bow Pod architecture allowing for efficient training of large numbers of such models, the possibilities for accelerating simulation workflows in HEP on the IPU are enormous.
In addition to the DiJetGAN model, the University of Bristol team implemented a recurrent neural network (RNN) for particle identification as well as a Kalman Filter algorithm on the IPU. More details can be found in their original paper, as well as in our blog.
These and other deep learning experiments in HEP have demonstrated the potential for AI to accurately model quantum interactions on a subatomic scale, but what about on an atomic or even molecular scale? Graphcore engineers have been working with researchers around the world looking to use AI to revolutionise fields such as quantum chemistry and simulating biological macromolecules.
We have seen how deep learning has the potential to revolutionise many aspects of simulation and modelling in sub-atomic physics, but there is also huge demand for fast and accurate simulation of microscopic objects on an atomic and molecular scale.
Molecular dynamics simulation is the simulation of movement in molecular and atomic systems, and typically involves calculating the energies of atoms in a molecule, the forces acting upon each atom, or both. Naturally, such computation quickly becomes extremely complex as the number of atoms considered increases.
Whilst the physical laws governing chemistry and a large portion of quantum physics are well-established, the respective equations are often too complicated to be solved exactly. Approximate numerical simulations often take a large amount of time, and so new approaches are required to accelerate these simulation tasks. Most recently, machine learning-based approaches have seen wider use in the development of tools for simulating complex atomic systems.
DP Technology, a leading Chinese organisation in molecular dynamics simulation for drug discovery, recently announced IPU support for their award-winning simulation platform DeePMD-Kit. This work, enabling more accurate simulation of molecular dynamics, orders of magnitude faster than with traditional numerical approaches, opens new possibilities in this field, which is currently undergoing an AI-driven transformation.
More information on this project can be found in our technical deep dive blog, as well as the DeePMD-Kit open source repository.
Several deep neural network-based approaches have recently been developed for molecular dynamics simulation in quantum chemistry.
SchNet is a GNN-based model developed for modelling quantum interactions between atoms in a molecule. Unlike pixels in images, the atoms in a molecule are not confined to a regular grid-like structure. Furthermore, their precise location forms crucial information necessary for calculating energies and inter-atomic forces. This model therefore makes use of continuous-filter convolutional layers so as not to require atoms’ locations to be discretised.
Using continuous-filter convolutions allows for the model to consider an arbitrary number of neighbouring atoms at arbitrary positions. The model consists of an embedding layer followed by three interaction blocks containing these continuous-filter convolutional layers, with interactions between atoms being computed by atom-wise dense layers, as shown in the image below. 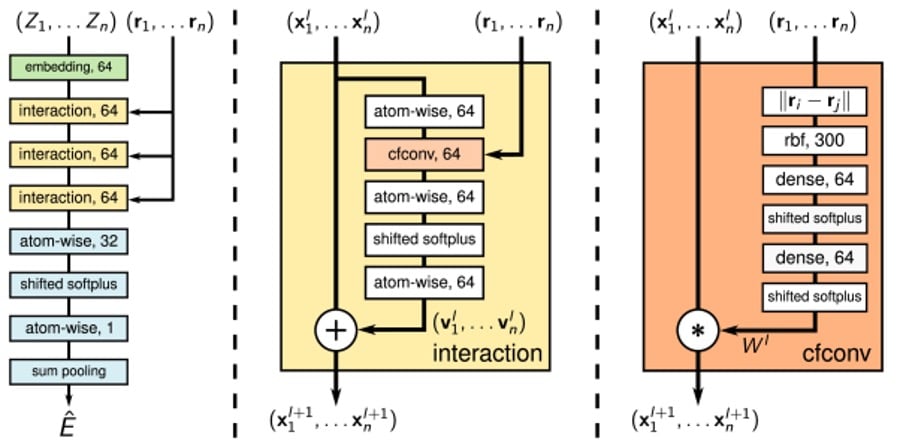
Figure 2: SchNet architecture, taken from ArXiv:1706.08566
Graphcore engineers successfully trained the SchNet model on IPUs on the QM9 molecules dataset, a dataset widely used to benchmark a model’s ability to predict various properties of molecules in equilibrium.
In addition to applications in quantum chemistry, deep learning has shown huge potential when it comes to protein-folding; the procedure by which a protein’s amino acid sequence is “folded” into its 3D atomic structure. Gaining better understanding of how a protein folds into its native 3D structure has been of much interest in computational biology for many decades, and the ability to accurately and efficiently model this behaviour would enable faster and more advanced medical drug discovery.
DeepDriveMD is a deep learning-driven molecular dynamics workflow for protein folding which combines machine learning techniques with atomistic molecular dynamics simulations. This hybrid HPC/AI approach consists of HPC-based numerical simulation and an AI surrogate model, presented in Figure 3.
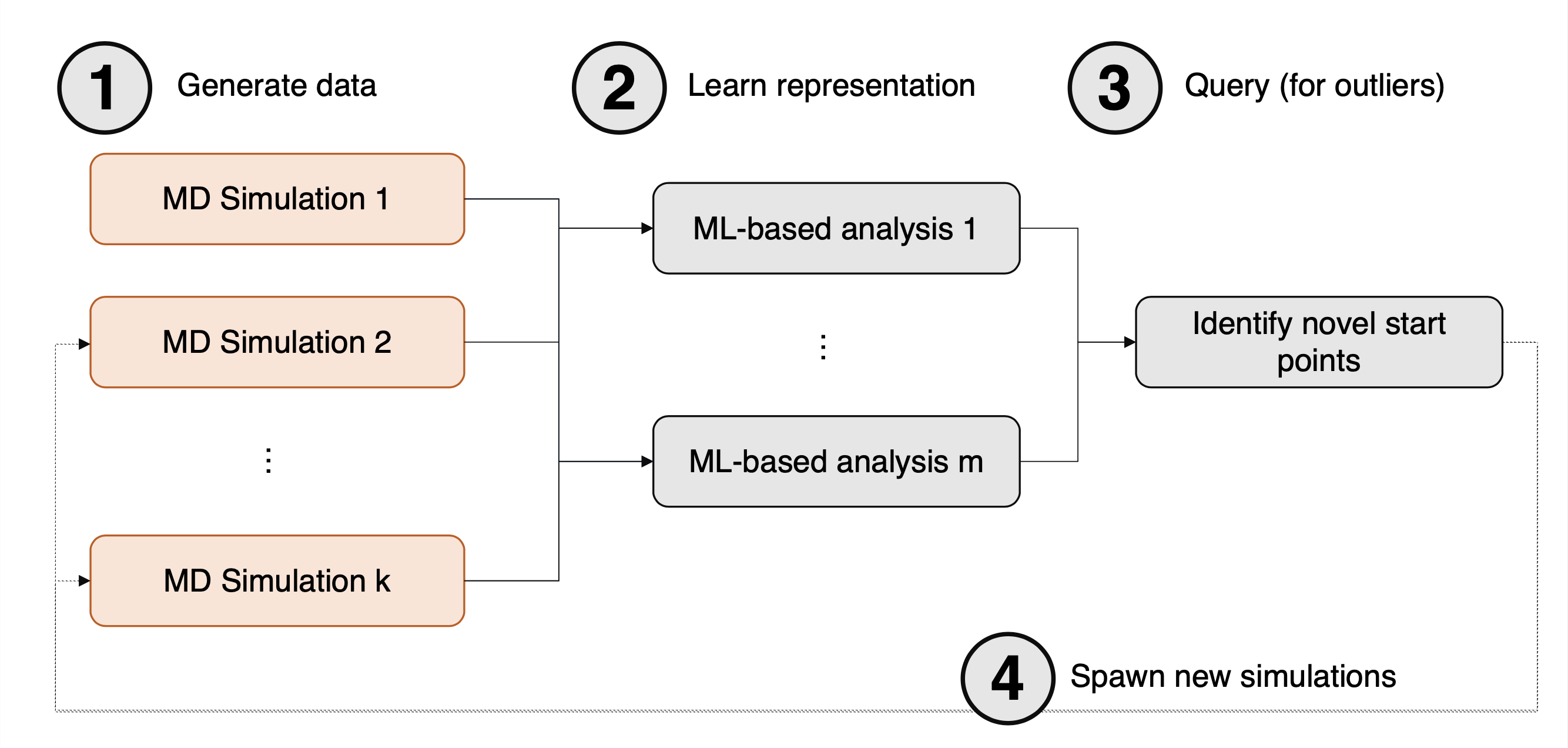
Figure 3: Computational motif detailing the 4 key steps of the DeepDriveMD toolchain, taken from ArXiv:1909.07817.
First, an initial pool of data is generated from a large number of MD simulations (1). Next, this data is fed as input into a machine learning model (2), followed by running inference on the model to identify new starting points for MD simulation (3). Finally, in (4) new MD simulations are generated. These can either simply be added to the pool of simulation data, or they can replace existing simulations; for example, simulations which have become stuck at some metastable state.
The machine learning model originally implemented as part of DeepDriveMD is a Convolutional Variational Autoencoder (CVAE), noting that the motif above is not restricted to a specific deep learning architecture. With this model trained, the authors were able to fold Fs-peptide folded states in 6µs compared to around 14µs without using an ML-driven approach, resulting in more than 2x speedup.
Cray Labs’ CVAE implementation of DeepDriveMD, part of Cray’s SmartSim repository, was trained on IPU-M2000 and throughput was around 3x faster than on an A100 GPU. Combined with the speedup achieved by using DeepDriveMD over a non-ML-based implementation, the combination of this hybrid AI/HPC approach and Graphcore IPU-PODs offers huge potential in accelerating protein folding workloads.
As we have seen with the recent use of neural networks in Computational Fluid Dynamics, AI-based solvers are becoming increasingly popular for computing good approximations to complex equations in an accelerated time frame. Another recent machine learning focus, which has been gaining popularity more recently, is Physics-Informed Neural Networks (PINNs).
In a typical supervised learning scenario, a neural network contains a loss function, which represents some measurement of how far away the network’s predictions are from the ground truth. Simple loss functions such as Root-Mean-Square Error (RMSE) simply use the difference between the predicted values and ground truth, without taking into account any prior knowledge of the physical system. In cases such as image classification such knowledge may not exist for arbitrary images, however in many areas of scientific research the underlying physics is often well-defined, for example by a set of differential equations. PINNs include these known equations as part of the loss function, making them better able to learn the behaviour of a particular system.
Researchers at Texas A&M University High Performance Research Computing (HPRC) have been investigating the use of PINNs to solve the infamous viscous Burgers’ Partial Differential Equation (PDE). This PDE occurs in various mathematical fields such as fluid mechanics and traffic flow and can be used to model wave evolution in incompressible fluids. The Burgers’ PDE has been studied by many researchers for over a century and is often used to test the accuracy of numerical PDE-solving programs.
An accurate, numerical approximation was used as a reference against which the PINN solution was measured. The PDE was constructed with sinusoidal initial condition and homogeneous Dirichlet boundary conditions as follows:
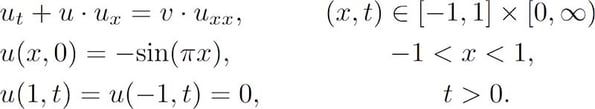
In the case where the fluid’s viscosity, v, is smaller than ~0.1π a discontinuous shock-wave forms at x=0.
The PINN solution of the viscous Burgers’ PDE was calculated using TensorDiffEq, an open-source TensorFlow 2.X-based package developed by researchers at Texas A&M University. This solution was found to be in excellent agreement with classic numerical solutions, with both solutions becoming unstable for very low viscosity, whilst taking less time to complete.
The viscous Burgers’ PDE implementation and the TensorDiffEq framework was run on IPUs, allowing for efficient acceleration of a multitude of PINNs.
We have seen several use cases where AI models are accelerating HPC workflows. And while the IPU has been designed to accelerate machine intelligence workloads, many aspects of the IPU architecture make it highly capable of performing very well for classical HPC workloads.
We have already seen the excellent results achieved by researchers at ECMWF using MLP and CNN models for weather forecasting applications, but there are also opportunities for the IPU to accelerate more traditional algorithmic approaches when it comes to climate modelling.
Graphcore engineers have been working with Chinese digital transformation specialists Agilor, modelling evapotranspiration; the rate at which water moves from surfaces such as plants and soil into the atmosphere. The aim in this case was not to run the entire end-to-end toolchain on the IPU, but to identify key elements which could be efficiently accelerated.
Measuring evapotranspiration can be incredibly useful for enabling precise irrigation in agriculture and is also being actively investigated for use in forest fire prevention and natural disaster management. There is, however, a limit to how spatially fine-grained such measurements can be in the real world, often resulting in measurements being quite coarse when plotted on a map. In order to give a finer-grained approximation of a location’s reference evapotranspiration value (ET0), an interpolation technique called Kriging is used. This method is closely related to regression analysis and is highly computationally intensive.
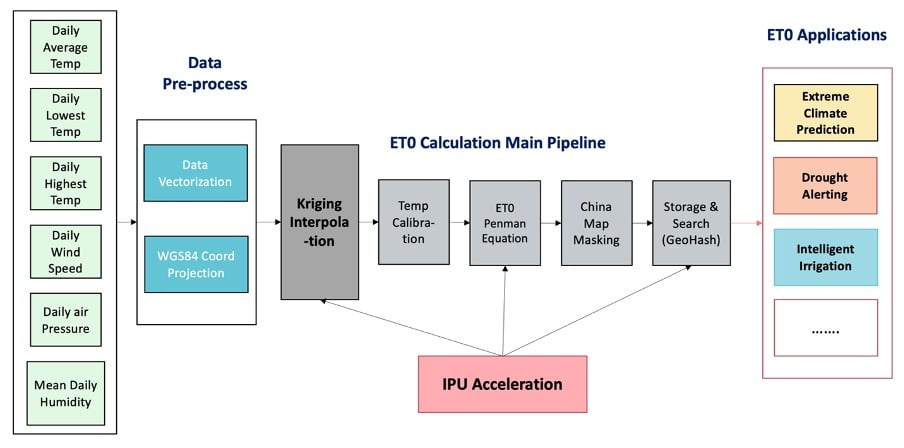
As shown in the figure above, the main calculation pipeline including the Kriging algorithm was run on IPU, keeping the data pre-processing and post-interpolation application steps on the CPU.
PyKrige, a Python implementation of the Kriging algorithm, was implemented in TensorFlow in order to perform the underlying matrix inversion and multiplication on the IPU. This enabled the interpolation of the entire dataset to be performed in just 21 seconds, compared with 2000 seconds using PyKrige on CPU.
More information on Agilor’s work on the IPU can be found in our technical blog.
While we have covered a wide range of exciting applications in the sphere of AI for Simulation, whereby traditional HPC workloads can be enhanced by AI techniques running on the IPU, this is just the tip of the iceberg. With large-scale scientific experiments such as CERN’s LHC looking to collect and analyse orders of magnitude more data in the coming years, the need to accelerate classical processes is greater than ever.
More and more researchers working in fields such as drug discovery, weather forecasting, climate modelling and computational fluid dynamics are looking towards ML-based approaches to enhance their toolchains, both in terms of accuracy and time-to-result. Furthermore, new approaches such as PINNs are revolutionising how neural networks can learn to emulate physical systems governed by well-defined yet complex equations.
With the ongoing development of these and other novel AI-based approaches, the need for specialised hardware capable of accelerating these workloads efficiently is growing. Graphcore’s IPU, designed from the ground up for machine intelligence, is the ideal platform on which to build, explore and grow the next generation of machine learning-driven solvers, emulators and simulations.
Sign up for Graphcore updates:
Mar 07, 2024 \ Research, Natural Language Processing, Generative AI, LLM

Feb 05, 2024 \ AI, Developer, Research

Dec 14, 2023 \ Research, Natural Language Processing, Large models, FP8, LLM

Sign up below to get the latest news and updates: