New Intelligence Processing Unit (IPU) hardware shows state of the art performance for next generation AI models which could translate to unprecedented accelerations for drug discovery applications.
Identifying effective new medicines is essential to the advancement of global healthcare. Yet the process itself of drug discovery is notoriously lengthy and costly, with many novel drugs and treatments failing to make it through clinical trials.
New machine intelligence approaches are helping to reduce the time and cost involved in drug discovery processes. By identifying patterns in large volumes of data, AI algorithms allow scientists to design new molecules, validate drug targets and even repurpose existing therapies for other medical applications.
Many emerging AI models that have the potential to enable further breakthroughs in drug discovery are slow to run due to the limitations of current hardware architecture. This is because GPUs are not built to efficiently support the sparsity and massive parallelism that we see in many of today’s newest models. Graphs, Probabilistic models and Massive Transformer models are all inherently sparse architectures that are either too slow or too costly to run on a GPU.
Here, we highlight key benchmarks including several innovative and relatively unexplored use cases for machine intelligence in healthcare.
New Hardware Architecture for Drug Discovery
Graphcore created the IPU as a fundamentally different processor architecture to GPUs and CPUs, designed specifically for machine intelligence. While GPUs incrementally improve from generation to generation, bringing their historical limitations with them, the IPU champions an entirely new compute workload. The IPU processor is not only optimized for today's most demanding machine learning workloads but has been built specifically for the complex computational demands of emerging models. Unlike GPUs, this new hardware architecture easily handles the massive parallelism, sparse data structures, low-precision compute, model parameter re-use and static graph structures that dominate next generation AI models.
So how does this novel architecture translate to state-of-the-art accelerations for next generation AI models used in the field of drug discovery? A number of companies and organizations working in the field of healthcare are already exploring how Graphcore's IPU can help advance their work, including Oxford Nanopore, Arzeda and Microsoft. Here are some of the machine intelligence approaches we are investigating that could drive huge advances in AI-powered drug discovery.
Multi-Layer Perceptrons
Multi-Layer Perceptrons are used in many different applications within genomics including phenotype classification, gene ontology prediction and genome sequencing.
Graphcore is currently working on a collaboration with Arzeda to model biological sequences, supporting the creation of novel proteins. By learning Markov Random Fields, we are able to describe protein families and folds, and then approximate this Markov Random Field as a supervised learning problem.
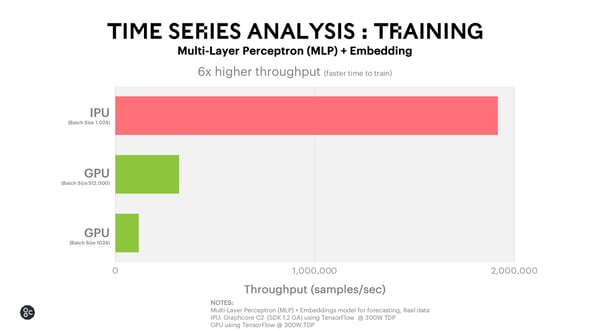
For this Multi-Layer Perceptron use case, the IPU enables 6x faster time to train.
Natural Language Processing
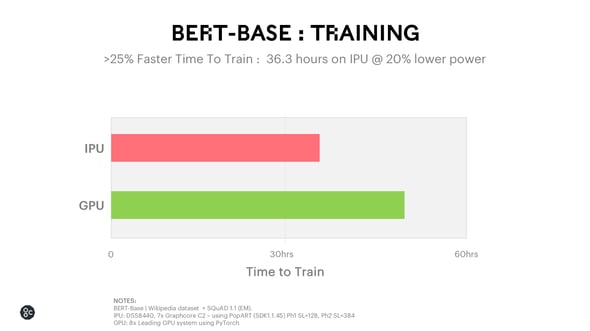
BERT is a family of Transformer models that have taken the NLP community by storm. These attention-based Transformer models deliver high levels of accuracy when analysing structure and meaning in text – an important factor for AI modelling in medicine. BERT can be fine-tuned for more specific types of language data if pre-trained on a particular corpus of terminology. For healthcare applications, the model can be trained on text from patient transcripts and medical literature. For BERT-BASE training, the IPU is able to achieve 25% faster training time at 20% lower power, meaning the algorithm will run faster at a lower cost.
IPU architecture is well suited to these types of emerging NLP models and efficiently runs innovations such as Block-Sparse based Transformer models, aided by the support of our static block sparsity Poplar software library.
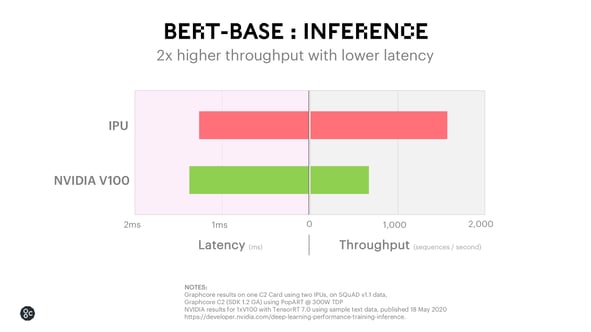
For BERT-BASE inference, the IPU provides 2x higher throughput, making it possible to use BERT in an interactive setting where scalability is a priority.
These benchmarks also generalize very naturally to genomics. The same NLP modelling techniques can be used for genome sequencing and to design new proteins more quickly and easily, which could reduce testing time in the lab.
Probabilistic Approaches
Our IPU hardware architecture lets innovators accelerate newer models. However, it also enables them to get the most out of existing models that, until now, have been limited by GPUs.
Markov Chain Monte Carlo (MCMC) is a good example of a classic machine learning algorithm that, though used across many different scientific domains, is perceived as slow. We believe that Bayesian algorithms such as MCMC are not actually inherently slow but simply do not run efficiently on a GPU.
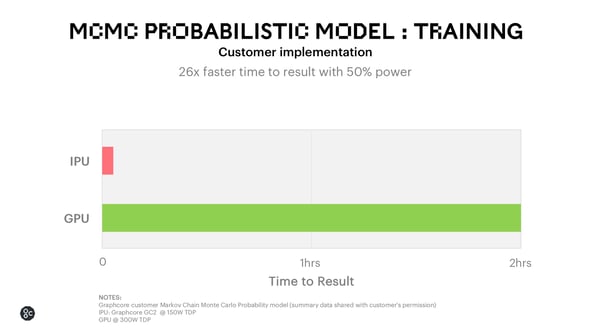
On the IPU, we see very compelling performance for MCMC algorithms. For a customer implementation we delivered 26x faster time to train for MCMC using 50% of the power when compared with a leading GPU.
In an “out-of-the-box” implementation using TensorFlow probability, the IPU still achieves 15x faster time to train.
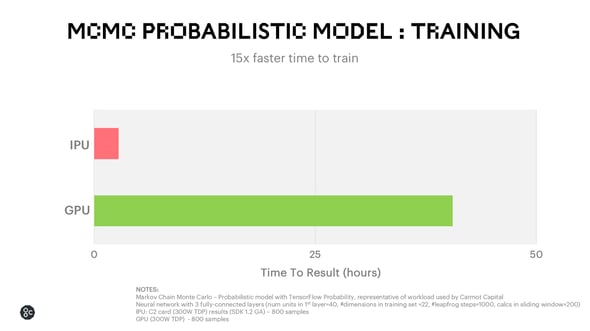
There is strong potential to apply this probabilistic model speedup to the field of drug discovery for tasks such as estimating the oral absorption rate of a drug or assessing the effectiveness of new pharmaceutical treatments.
Computer Vision
EfficientNets were designed using neural architecture search to obtain higher accuracy and efficiency than previous Convolutional Neural Networks. Because of this improved accuracy, this family of models can be applied very effectively to computer vision tasks, including diagnostic imaging.
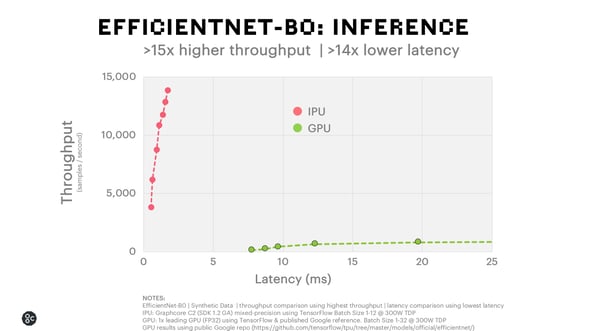
When running EfficientNet-B0 on the Graphcore IPU, we can deliver 15x higher throughput and 14x lower latency for inference and achieve 7x faster time to train than the leading GPU alternative.
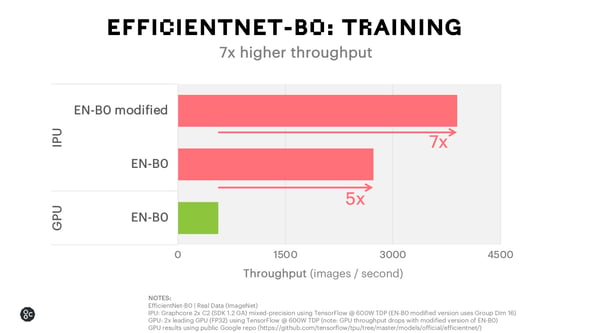
We have also been working with Microsoft to explore potential accelerations of new computer vision models on the IPU. For EfficientNet-B0 on the IPU, Microsoft researchers completed training runs in 40 minutes using the Graphcore IPU on Azure compared to 3 hours on a high-end GPU system.
The data used for these training runs was a set of pneumonia chest X-ray images from RSNA‘s 2018 Kaggle challenge, combined with a smaller set of publicly available Covid-19 X-rays to test the system’s applicability to the novel coronavirus. Such a significant acceleration makes it much easier to envisage applying EfficientNet training to diagnostic imaging in real-world medical situations, where timing is critical.
Future Directions for AI-Powered Drug Discovery
New hardware architecture lets scientists look beyond what is currently possible in drug discovery to use new and previously unexplored AI approaches. In the case of MCMC, we have seen that even traditional machine learning algorithms can be accelerated with an architecture designed specifically for AI workloads.
We look forward to seeing the advances in medicine that will be made possible by the IPU as innovators begin to leverage next generation models and accelerate their AI workloads.
Graphcore is always open to collaborations with real-world practitioners and we will report on results from our current exploratory work with organizations such as Oxford Nanopore, Arzeda and Microsoft as these projects progress.
