
Jul 05, 2023
Graphium: an IPU-ready Python Library for training molecular GNNs at scale
Written By:
Dominique Beaini

Jul 05, 2023
Written By:
Dominique Beaini
We're Hiring
Join us and build the next generation AI stack - including silicon, hardware and software - the worldwide standard for AI compute
Join our teamGraphium is an open-source library designed for graph representation learning on real-world chemistry tasks.
Graphium provides access to state-of-the-art GNN architectures via an extensible API, enabling researchers to build and train their own large-scale GNNs with ease.
From launch, Graphium is able to take advantage of the Graphcore IPU’s unique architecture, which has been shown by both commercial and scientific research organisatons to dramatically accelerate GNN workloads.
Users can experience Graphium's capabilities using the ToyMix dataset, running in a Paperspace Notebook. For more information on this notebook see our dedicated blog.
In recent years, there has been a significant surge in the adoption of graph representation learning, accompanied by the widespread use of graph neural networks (GNNs) across diverse applications ranging from social networks to roadmaps to drug discovery and beyond.
Representing molecules as graphs has grown in popularity given the potential of being able to better capture the complex nature of molecules. In this approach, atoms are represented as nodes and covalent bonds as edges, which can be used for downstream tasks such as property prediction, chemical reaction prediction, and molecular generation. As a result, GNNs have emerged as a leading architecture for learning these molecular representations.
To train machine learning models on molecular graphs, the conventional approach involves extracting a binary fingerprint vector based on local structural information of the graph that is then fed into the model. However, these approaches suffer from inherent limitations, such as bit collisions when processing large datasets and a strong inductive bias that assumes molecular properties depend solely on local structures.
With the recent advances of deep learning in complex tasks like computer vision and natural language processing, a natural next step is to adopt similar algorithms to molecular graphs.
By embracing this approach for state-of-the-art (SOTA) GNNs, we aim to replace pre-defined fingerprints with learned graph embeddings that can adapt to different tasks. This learned representation effectively eliminates the inductive bias of local structures.
Ultra-large, multi-layer GNNs could change the way we work with molecules as they can learn features beyond the graph connectivity that embed not only their topological structure but also their quantum mechanical and biological properties.
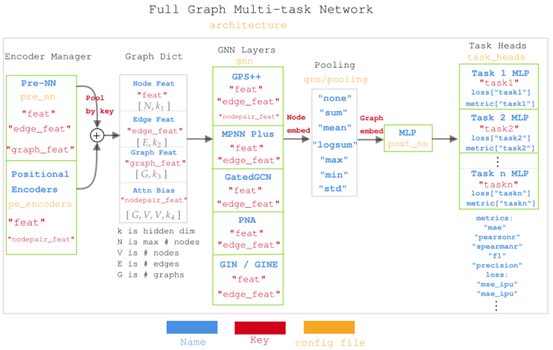
Recent advances in the field have reshaped our understanding of GNN design and emphasize the significance of various components. These include the types of positional encodings, the message passing layer, the edge-aware attention in Transformers, the movement of features across levels (nodes, edges, graphs, node pairs), and the ability to make predictions at each level.
This is where Graphium truly excels: it incorporates all of these advances in a flexible manner, enabling efficient exploration of the design space for GNNs and unprecedented speed, scale, and simplicity in graph-based workflows.
Graphium is an open-source Python library focused on graph representation learning for molecules at scale. With a built-in, best-in-class atom/bond featurizers and a modular and configurable API, Graphium makes it easy for anyone, regardless of past expertise in graph learning, to train molecular GNNs for applications in molecular property prediction, optimization, and design. By optimizing the library for multiple types of hardware, and implementing multi-level multitasking, Graphium is helping researchers worldwide unlock the true potential of GNNs.
Graphium was created by and for researchers who live at the intersection of graph machine learning and drug discovery. It is the only library dedicated specifically to training GNNs on molecular graphs at scale, including:
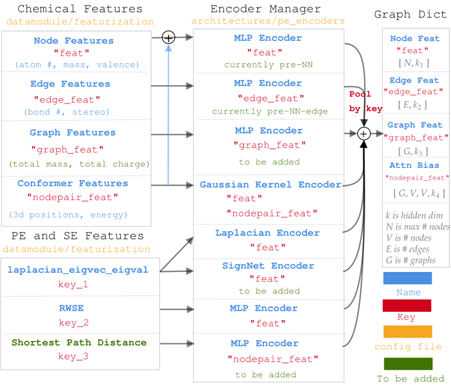
🤑 Rich Featurization: Graphium has powerful and flexible built-in molecular featurizers for property prediction. Instead of assuming a predefined set of molecular features, Graphium users can input what they deem to be most important for a given task, with 38 atomic features (mass, electronegativity, atomic number, group, period, etc) and 7 bond features (stereo, conjugation, length, etc). Conformational properties are also supported with the ability to process 3D positions using Gaussian kernels or other priors.
🎨 Powerful: Graphium makes it simple to implement the best and most recent GNN models via a configuration file with the necessary flexibility for research. Users can access both baseline and SOTA 2D and 3D GNN architectures. These include GCN, GIN, Gated-GCN, PNA, DimeNet, Graphormer, and the 2022 OGB-LSC award-winning model, GPS++.
🕹️ Easy to Use: With an extensible API, researchers can effortlessly build and train their own GNN models. Using a configuration file, users can define all parameters of the model and the trainer.
📖 Flexible: By keeping GNN layers independent of the model architecture, Graphium offers unparalleled code flexibility, enabling seamless swapping of different layer types as hyperparameters. Users can subsequently build full GNNs using any kind of GNN layer. For example, swapping your baseline GCN with GPS++ is as easy as changing a few lines in a YAML file. This increased flexibility enables Graphium to grow and evolve with the industry by continuously and efficiently integrating novel architectures, GNN layers, or pooling layers. It further allows practitioners to cross-validate parameters by decoupling the effects of these individual choices.
⚡ Fast: Based on the PyTorch-Lightning library, we provide a simple training loop that seamlessly handles the dataset, device type, and metrics logging. All the training parameters (learning rate, number of epochs, early stopping) and the metrics to track are configurable in a YAML file. The best model is saved automatically and ready to load for an inference task or for transfer learning. Graphium also offers ultra-fast training of GNNs with optimized data processing, data loading, and support for IPU accelerators, in addition to GPUs. IPUs facilitate faster sparse-matrix multiplications and enable faster and more cost-effective GNN training.
🔥 Scale: With an accompanying dataset of >86M unique molecules and up to 3.3k labels per molecule, Graphium offers massive, multitask supervised molecular datasets that are orders of magnitude larger than any other publicly available dataset. By training simultaneously on millions of molecules and thousands of tasks, users can train models that better generalize across the chemical space and tasks.
🦜 Expressivity: Positional encodings are an active area of research for GNN expressivity and are especially important for graph Transformers. Graphium supports more positional encodings than any other library, including eigenvectors, random walks, commute distance, heat kernels, and graph distances. Moreover, all of these positional encodings are supported at the graph, node, edge, and pairwise level due to the novel ability to transfer from one level to the other via pooling/unpooling. All positional encodings can be passed to specifically designed encoders, such as MLPs, SignNet, and Gaussian kernels before being appended to the graph features. With one library, you can better navigate the space of expressive GNNs and Transformers.
In addition to the code below, developers can also try our Graphium using the ToyMix dataset on Paperspace.
First, from the software download center, download the right Poplar SDK for your operating system. Then follow the installation instructions below.
mkdir ~/.venv # Create the folder for the environment
python3 -m venv ~/.venv/graphium_ipu # Create the environment
source ~/.venv/graphium_ipu/bin/activate # Activate the environment
# Installing the poptorch SDK. Make sure to change the path
pip install PATH_TO_SDK/poptorch-3.2.0+109946_bb50ce43ab_ubuntu_20_04-cp38-cp38-linux_x86_64.whl
# Activate popular SDK.
source PATH_TO_SDK/enable
# Install the IPU specific and graphium requirements
PACKAGE_NAME=pytorch pip install -r requirements_ipu.txt
pip install -r lightning.txt
# Install Graphium in dev mode
pip install --no-deps -e .
To learn how to train a model, we invite you to look at the documentation or you can also work with readily available models on Paperspace.
We believe GNNs can have a broader impact by extending their domain of applicability to other tasks in drug discovery. As we expand Graphium, we hope to unlock new tasks enabled by unprecedented scale and speed, such as advances in ultra-large docking, molecular dynamics, and protein design.
Follow datamol.io on Twitter to stay up to date on future announcements.
You can check out our documentation here or try Graphium online. We welcome your feedback on the Github repository, or the forum. You can also join the Learning on Graphs and Geometry (LoGG) Slack community where users can share their work, ask questions, and collaborate on projects using Graphium.
The Graphium library was created in collaboration between Valence Discovery, Graphcore and academics at MILA. The Graphcore team includes Alex Cunha, Blazej Banaszewski, Zhiyi Li, Sam Maddrell-Mander, Chad Martin, Dominic Masters, and Callum Mclean.
This blog was written by Dominique Beaini, Adjunct Professor at the University of Montreal in the Department of Informatics and Operational Research. He is also an Associate Industry Member at Mila, and is a Staff Scientist focused on graph research at Valence (recently acquired by Recursion).
Sign up for Graphcore updates: