Researchers from the University of Massachusetts (UMass) Amherst, Facebook and Graphcore have published a new paper demonstrating how important COVID-19 analysis using Approximate Bayesian Computation can be massively accelerated with IPU processors. The results indicate an impressive 30x speedup on IPUs compared with CPUs and a significant 7.5x speedup compared with GPUs.
At Graphcore, we’ve always spoken about accelerating innovation, but never has that idea felt more real than in 2020. As a society, we all benefit when scientists and researchers are able to speed up the computational element of their work. The sense of urgency around discovery is something we all now feel keenly.
As many of us now know, to tackle a pandemic effectively, it is important to understand how infections spread in a population and how different interventions can impact the spread. Parameters which are particularly useful for this purpose include the infection rate, recovery rate, positive test rate, fatality rate, and testing protocol effectiveness. Since the spread of a virus is not a deterministic process at a macroscopic level, researchers are interested in the distribution of these parameters rather than point estimates. Knowing these distributions enables us to find out where significant differences can be observed in contrast to spurious deviations.
Sourabh Kulkarni, UMass Amherst, presents the paper’s findings at the IEEE ICRC 2020 Conference
Two aspects make this modelling challenging. There is currently no “perfect” or “correct” model to describe the spread of the virus and some key figures are unknown. For example, we do not know the number of people who are infected but did not get tested or had a false-negative test, or the number of people who never got tested but recovered or died.
The field of simulation-based inference addresses the problem of such unobserved variables with an algorithm called Approximate Bayesian Computation (ABC). ABC is usually run on large numbers of CPUs. As there is currently no one “correct” model, fast computation is required to ensure that researchers can quickly iterate over different modelling approaches to determine which methods are the most appropriate. In our joint research, we have shown that the IPU can significantly accelerate the ABC algorithm, allowing us to more rapidly analyze and interpret the epidemiological patterns behind the COVID-19 outbreak.
Parallel ABC Inference on the IPU
The statistical COVID-19 transmission model is depicted in Figure 1 and the main ABC algorithm is described in detail in the paper: Accelerating Simulation-based Inference with Emerging AI Hardware, S Kulkarni, A Tsyplikhin, MM Krell, and CA Moritz, IEEE International Conference on Rebooting Computing (ICRC), 2020. A subsequent version of the paper, published in ACM Journal on Emerging Technologies in Computing Systems, can be found here. The code is open source and shared in Graphcore’s GitHub portfolio repository.
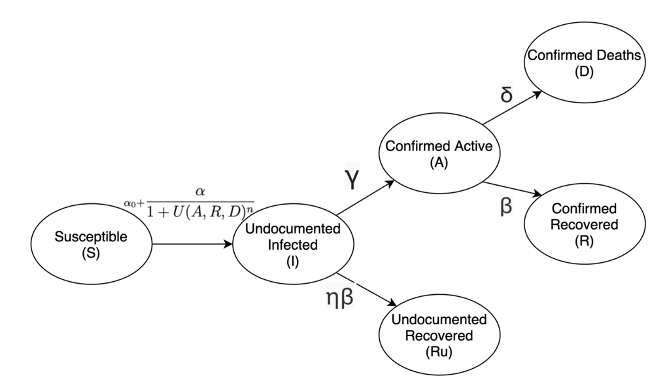 Figure 1: Covid-19 Transmission Model, as shown in the paper "Accelerating Simulation-based Inference with Emerging AI Hardware"
Figure 1: Covid-19 Transmission Model, as shown in the paper "Accelerating Simulation-based Inference with Emerging AI Hardware"
In short, we run 200,000 simulations in parallel on a single C2 card with two MK1 chips and 1.6 million simulations in parallel on a whole server with 16 IPUs. Each simulation plays out a different randomly sampled set of parameters. We then obtain an accurate distribution of model parameters by selecting only those that generate observations within a certain threshold of the real-world observations. Lowering this threshold improves the model fit, but also leads to an increase in computation required. Figure 2 provides such an example of parameter distributions, comparing the recovery rate of New Zealand and the USA at the first 49 days after the first 100 discovered cases.
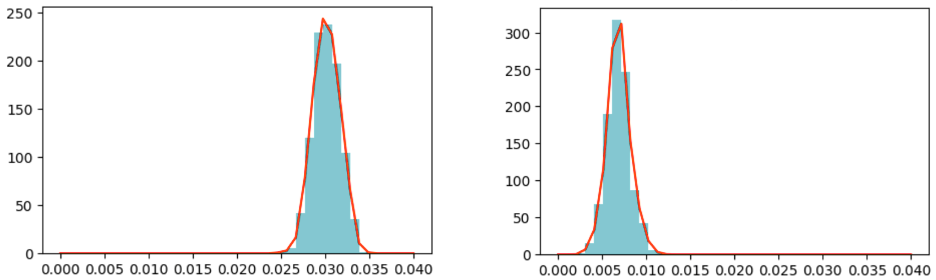
Figure 2: Parameter distributions of the estimated recovery rate of New Zealand [left] and the USA [right].
This parallelization accelerates the processing by a factor of 30 when comparing Graphcore’s C2 Card (2 MK1 IPUs) with the performance of two Xeon Gold 6248 CPUs which have the same Thermal Design Power (TDP) of 300W. The IPU is also faster than a Nvidia Tesla V100 by a factor of 7.5, which is also power-equivalent. This hardware was chosen due to its availability to the researchers at the time of the experiments.
What drives these accelerations?
In a more detailed follow-up analysis, our findings suggest three reasons which explain these speed-ups:
- The data arrays in the simulation are very large and do not fit entirely into the GPU cache. This means that the GPU cannot benefit from large batch sizes and requires a lot of rather slow communication to the main memory which is barely occupied by the data. Conversely, the MK1 IPU keeps all data on its larger on-chip memory (300MB) and benefits from increasing the batch size to its memory limits.
- The GPU requires a considerable amount of time to run the simulations, because it fetches the instructions from memory to the accelerator chip, a process known as ‘kernel launch’. In contrast, instructions reside in the IPU memory so no transfer of instruction code is needed and they occupy less than 40MB.
- Profiling using Graphcore’s PopVision Graph Analyzer tool shows that the simulation requires a lot of data communication. The IPU does not just have 50 times higher on-chip memory bandwidth compared to the GPU, but also more than 4 times greater compute throughput.
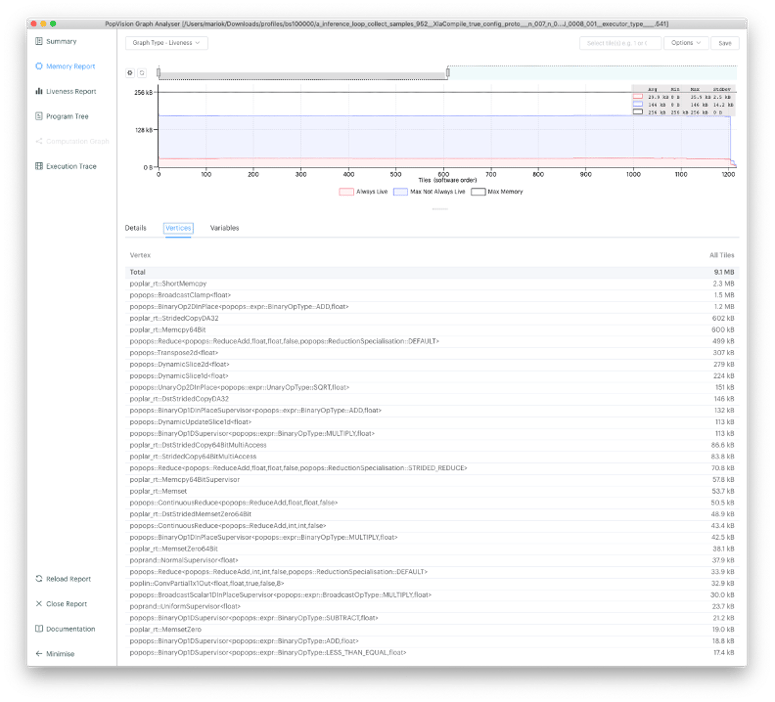 Figure 3: From our PopVision Graph Analyzer tool, it can be seen that a significant amount of memory is occupied by solely communication and data rearrangement operations on the IPU.
Figure 3: From our PopVision Graph Analyzer tool, it can be seen that a significant amount of memory is occupied by solely communication and data rearrangement operations on the IPU.
Future Applications
This unique approach to memory and data access leads to much faster computations on the IPUs when compared with GPUs. We hope that this acceleration will benefit COVID-19 modelling research and we are looking forward to seeing further applications of this nature in future, such as modelling the inter-regional spread of infections based on transit information, for example. This research could also be generalized to other statistical models that require simulation-based inference, and towards developing parallelized versions of simulation-based inference algorithms other than ABC.
Read the full paper (a subsequent version, published in ACM Journal on Emerging Technologies in Computing Systems, can be found here)
Review the Code
